 Sara Bell joined the program on January 1, 2020, while a second-year student, and is expected to graduate in 2023. Her graduate program is chemistry, and she completed her undergraduate degree in chemistry at Illinois State University. Her current research is in developing small-volume mass spectrometry-based approaches to characterize neurochemical dynamics in rodent and fish brains.
Sara Bell joined the program on January 1, 2020, while a second-year student, and is expected to graduate in 2023. Her graduate program is chemistry, and she completed her undergraduate degree in chemistry at Illinois State University. Her current research is in developing small-volume mass spectrometry-based approaches to characterize neurochemical dynamics in rodent and fish brains.
In March 2020, she presented her research on “Off-Line Matrix Assisted Laser Desorption/Ionization-Mass Spectrometry of Microfluidic Droplets for Neurochemical Detection” at the Pittsburgh Conference on Analytical Chemistry and Applied Spectroscopy in Chicago. Her research also contributed to a poster at the June 2020 ASMS Conference on Mass Spectrometry and Allied Topics. Additionally, she has presented her research at the 2020 and 2021 MBM Retreats. Her first-author journal article “Droplet Microfluidics with MALDI-MS Detection: The Effects of Oil Phases in GABA Analysis” was published in ACS Measurement Science Au in August 2021. Bell was also a co-author on the article “Single Cell Classification Using Mass Spectrometry Through Interpretable Machine Learning” published in Analytical Chemistry in 2020. She recently (2021) submitted for review a co-first-author review to Annual Review of Analytical Chemistry regarding neuropeptide detection by mass spectrometry. Currently, she is completing lab rotations under the advisement of Jonathan Sweedler and Rashid Bashir. She began a collaboration with Alison Bell’s lab in September 2020 as her external research experience, with the goal of profiling single cells from three-spined stickleback fish to elucidate connections between peptide profiles and parental behaviors.
Bell has completed three semesters of the Special Topics in MBM course and participated in other trainee activities such as the Summer Journal Club readings/discussion meetings and Frontiers in Miniature Brain Machinery lectures. In spring 2021, she developed a short video tutorial on Single Cell Mass Spectrometry for the Beckman Virtual Open House. Other community outreach activities include serving as a Women Chemists Committee Mentor to women-identifying first-year graduate students (Fall 2019 to Spring 2020, Fall 2021 to Spring 2022) and as a representative on the Department of Chemistry Graduate Student Advisory Committee to plan and coordinate professional development opportunities (2020 through 2021). In addition, she has mentored a graduate student in the Jonathan Sweedler Lab. She joined the MBM Student Leadership council in January 2021.
Bell has completed coursework in integrative neuroscience, evolutionary neuroscience, and professional development for chemists (modules in teaching, professional ethics, & non-technical skills) in addition to advanced chemistry coursework. In January 2020 she attended a Responsible Conduct of Research Seminar and in February 2020 attended “Teamwork, Communication, and Research Integrity.” In June 2020 she attended seminars on “MS Career Options: How to Kick Start Your Career,” and “Make Today Count for Tomorrow: Virtual Professional Development Lunch.” In January 2021 she attended the Reach Out Science Communication Challenge virtual workshops hosted by NSF and the Museum of Science in Boston. In April 2021, she was a finalist in the University of Illinois Research Live! Showcase for graduate student presentations. In July 2021, she participated in an American Chemical Society professional development workshop on improving science outreach and safety culture.
Research Highlights (in her own words):
My research focuses on development of bioanalytical MS methods for interpretation of the dynamic chemical profiles of single cells and brain extracellular fluid collected by droplet-nanodialysis. Specifically, I developed methods to use matrix assisted laser desorption ionization – mass spectrometry (MALDI-MS) to perform high throughput analyses. These methods are used to profile small molecules (<500 Da) in artificial cerebral spinal fluid droplets and peptides from the pituitaries of three-spined stickleback fish, gasterosteus aculeatus.
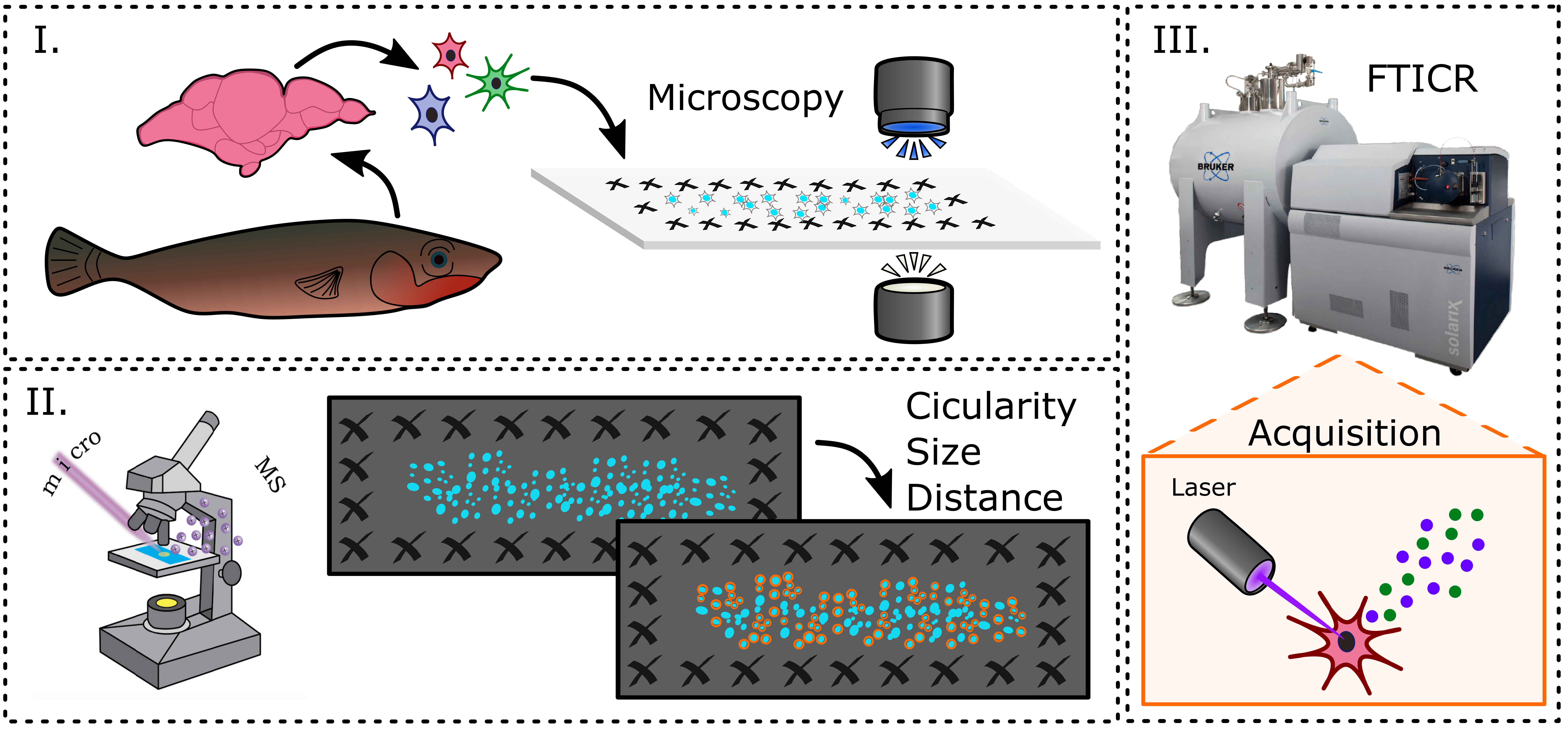
My main project is a collaboration with the labs of Professor Yurii Vlasov and Professor Rashid Bashir. Our goal is to collect release of small neurochemicals (<500 Da) within the cortex extracellular space by a droplet microfluidic, silicon nano-dialysis probe for analysis by MALDI-MS. The probe tip is 15 x 70 um in order to decrease damage to the surrounding brain tissue and to increase spatial resolution of neurochemical capture. Previously, I have worked with droplets generated from standard solutions using a PDMS device capable of producing volumes down to 65 pL. In these studies, we determined an optimal oil to use for droplet generation that did not interfere with MALDI-MS detection of the small molecule neurotransmitter GABA. From these studies we were able to produce a droplet generation-MS detection pipeline which was recently published in ACS Measurement Science Au. Currently, we are applying this pipeline to generation and analysis of pL droplets using the silicon probe produced by the Vlasov lab.
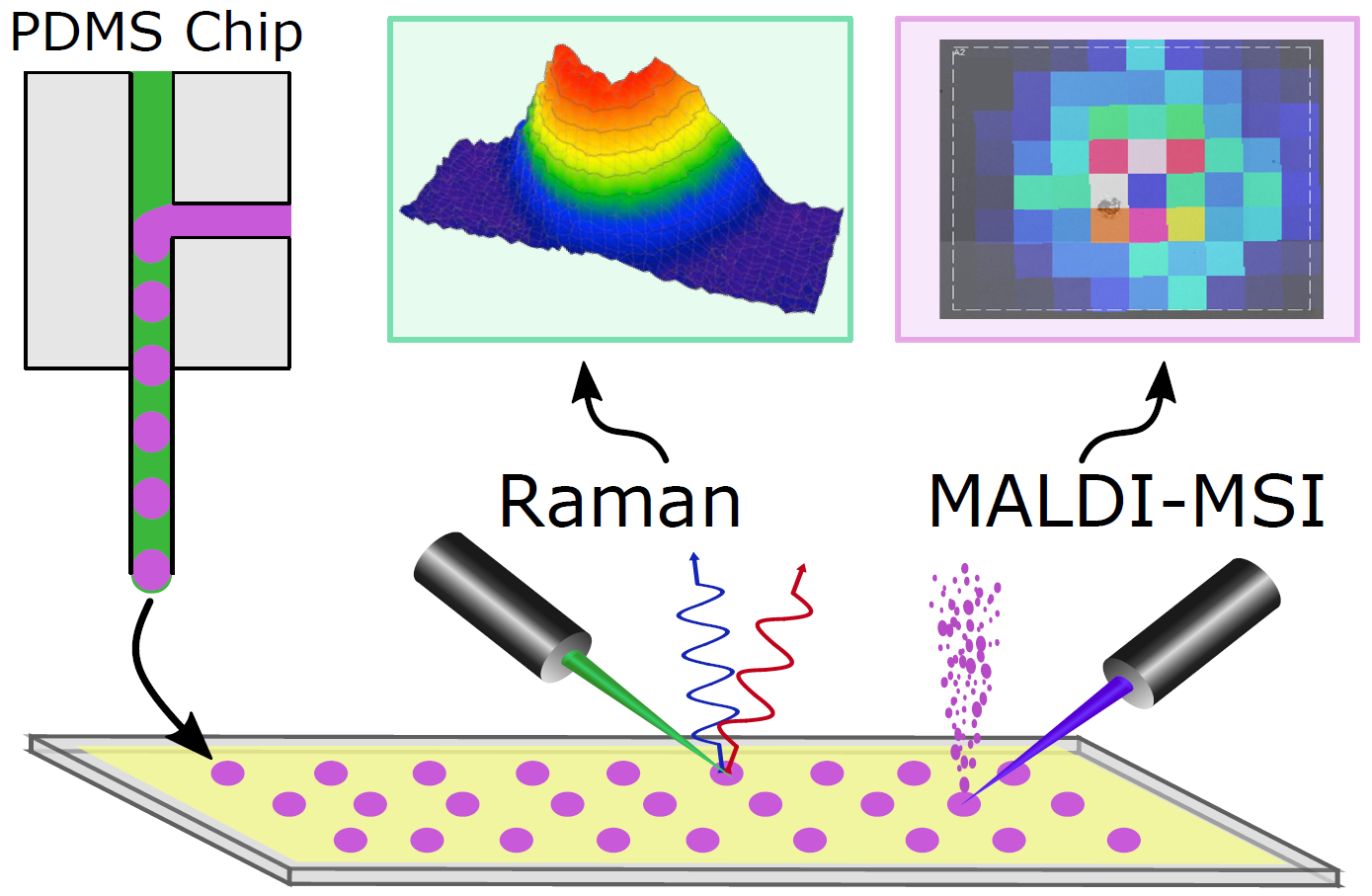
Before we can begin any in-vivo studies, we need to optimize transfer of droplets from the Si probe to a MALDI-MS compatible substrate and determine our MS detection capabilities. The Si probe produces droplets between 5 and 40 pL containing a fluorophore for visualization. Due to the small size of the tip and small volume of the droplets, the droplets must be removed from the probe by using some force. In this case, application of voltage to the probe tip and the substrate creates a potential difference moving the drops from the tip to the substrate. After my Vlasov lab collaborators deposit droplets on the substrate, I use fluorescence and brightfield microscopy to determine the location of the droplet deposits. This image is then used to guide MS imaging of the area. The MS images of the droplets and surrounding area are used to determine 1) whether droplets were successfully deposited on the substrate and 2) whether the MALDI-MS signal is co-localized with the droplet deposits seen in optical microscopy.
Our results using 40 pL droplets were recently presented at the 2021 microTAS conference. In this case, 41 droplets containing 8 fmol of GABA were imaged and the MS spectra corresponding to each droplet were compiled for statistical analysis of many parameters including standard deviation of GABA signal intensity, localization of GABA signal to the droplet deposit, and an estimated limit of detection below 100 amol. This type of analysis has led us to conclude that the GABA signal is well localized to visible fluorescent deposits corresponding to droplets. This serves as the basis for our current efforts using 5 – 10 pL droplets containing 7 neurochemicals at amoles/droplet each.
Additionally, I am continuing a collaboration with Professor Alison Bell, as part of the MBM research exchange, with the goal of profiling single cells from three-spined stickleback fish to elucidate connections between peptide profiles and parental behaviors. Male stickleback are the caregivers for their young and perform well characterized tasks from the time eggs are laid to a week post hatching. The Bell lab has established that distinct differences in paternal behaviors are common within and across populations. Furthermore, they’ve shown that plasticity and variation in paternal behaviors are heritable. Therefore, it is worth investigating the genetic connections to these behaviors. The Bell lab is interested in understanding genetic differences between two populations of stickleback fish that exhibit very different parenting styles, the attentive Putah fish and the un-attentive white fish. To investigate the biochemical reasons behind these differences and to compliment the Bell lab’s single cell transcriptomics data, I performed single cell analysis of the pituitaries from Putah and white fish collected during their breeding season.
As preparation for this task, I worked on optimization of fish single cell MALDI-MS through the use of pituitary cells. We have been using the pituitary since it is highly peptidergic and the peptide content is well preserved across species. Therefore, we expect to be able to see peptides of known identity in the MS results. I worked with Dr. Stanislav Rubakhin in the Sweedler lab to determine the best procedure for dissociation of single cells and MALDI preparation. After developing a procedure which gave reproducible and strong detection of expected pituitary peptides, this method was applied to pituitaries from Putah and white fish taken during the summer breeding season. MS data for ~40,000 single cells were collected using high mass resolution Fourier Transform Ion Cyclotron Resonance (FTICR)-MS for machine learning analysis of the peptide and lipid contents. We are currently performing this analysis to determine which peptides are different between the two fish populations. Finally, mass signals will be matched to an in-house, LC-MS peptide library and the resulting protein/peptide families that are found will be compared to genetic data from the Bell lab.
Reflections on MBM:
The MBM program has provided many opportunities to share my research, hear from other researchers, and learn helpful professional development skills. First, I participated in the Reach Out Science Communication Workshops held by the National Science Foundation and the Boston Museum of Science. This was a great opportunity to learn better techniques for presenting research to non-technical audiences and provided exposure to talks from other research areas. These workshops were very helpful when I prepared a short video explaining single cell analysis for the MBM’s contribution to the Beckman Virtual Open House. Additionally, I used the skills learned there to prepare my Research Live presentation and qualified as a finalist. I thoroughly enjoyed the Research Live experience as it was great to get feedback from judges outside of my research area and it was interesting to hear about the other fascinating research taking place on campus. Within the MBM class, I’ve enjoyed hearing about the research done both here at UIUC and other universities. Furthermore, I’ve appreciated the multiple talks given by representatives of The Career Center. Overall, I benefited greatly from these opportunities available through the MBM.

